SeqGI - Sequence Read Enrichment at
Genomic Intervals - is a
collection of scripts that can be used to analise and visualise
genome-wide ChIP sequencing output data. All scripts are
written in Python and R.
SeqGI can be used for common research questions faced at the initial steps of the analysis, for example:
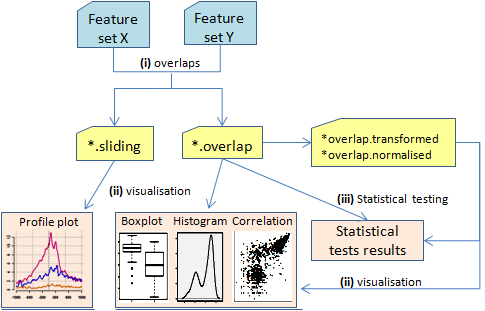
SeqGI workflow
The initial step in a workflow within SeqGI is the computation of overlaps between two feature sets. In a typical ChIP-Seq or RNA-Seq analysis flow, SeqGI can be used to quantify the ChIP/RNA signal at genes (or other genomic features of interest).
SeqGI can be used for common research questions faced at the initial steps of the analysis, for example:
- What are the number of reads overlapping a list of promoters?
- Are reads enriched at a particular group of promoters?
- What is the distribution pattern of a given epigenetic mark around a given genomic feature of interest (such as genes TSSs)?
- What is the statistical significance of differential ChIP-Seq signals between groups of genes or between samples?
SeqGI workflow
The initial step in a workflow within SeqGI is the computation of overlaps between two feature sets. In a typical ChIP-Seq or RNA-Seq analysis flow, SeqGI can be used to quantify the ChIP/RNA signal at genes (or other genomic features of interest).

Scripts description
(i) OVERLAPS:
The initial step in a workflow within SeqGI is the computation of overlaps between two feature sets.
1) Go to the Download page, download the latest zip files (SeqGI_V1.2.zip and testfiles.zip)
2) To let you started right away, we included some test files (testfiles.zip), just follow the Quick Start Guide and you can start testing SeqGI right right away!
3) We recommend that you go through the content of this webpage (OGRe usage and GenomeIntervals2BED). You can find several interesting examples that might inspire you and that you might want to do!
(i) OVERLAPS:
The initial step in a workflow within SeqGI is the computation of overlaps between two feature sets.
- OGRe script is a python
script that can be used to intersect a
BED, BedGraph or output files from standard aligners with a set of
dictated genomic features.
- The overlap between two genomic features
(e.g. reads overlapping genes) can be quantified using two metrics:
- By considering the number of features
- By considering the depth of coverage (also known as the breadh of the overlap)
- GenomeIntervals2BED.py
is a python script that can be used to convert a text file with
chromosome, start, end in any order into a standard bed file. In
addition, it can be used to specify gene regions of interest, for
example:
- 2kb upstream TSS, 1kb downstream TESs, 500bps promoters centered at the TSS
- or any region relative to gene positions!
- SeqGIplots_sliding.r can be used to visualise read coverage across genomic intervals as average profiles;
- SeqGIplots_overlap.r can be used to visualise the distribution of read coverage using boxplots or histograms. Correlation plots can also be used to correlate two samples.
- Check out the Quick Start Guide section for examples!
- Data normalisation, transformation, filtering and a variety of statistical tests that can be used to compare groups within one condition (one sample), or to compare conditions (two samples);
- Normalisation options include quantile normalisation, RPKM, median scaling, and others;
- Classical parametric (e.g. t-test) and non-parametric (e.g. Wilcoxon rank test) tests can be used to compare groups within a given condition (e.g. compare different groups of genes/peaks/intervals within one sample);
- The methodology implemented in several Bioconductor packages (such as DESeq, DEGSeq, edgeR, and others) can be used with SeqGI's count-type data to test the differential read enrichment between samples (e.g. compare a group of genes/peaks/intervals between wild-type and knockdown; or undifferentiated and differentiated conditions, etc).
1) Go to the Download page, download the latest zip files (SeqGI_V1.2.zip and testfiles.zip)
2) To let you started right away, we included some test files (testfiles.zip), just follow the Quick Start Guide and you can start testing SeqGI right right away!
3) We recommend that you go through the content of this webpage (OGRe usage and GenomeIntervals2BED). You can find several interesting examples that might inspire you and that you might want to do!
