Table of contents
1. Overview
2. Option summary
3. Example usage
5. Output file format (BED)
2. Option summary
3. Example usage
3.1. Convert any (yes! ANY) text file to BED: default
behaviour (--window=None)
3.2. Define a window relative to the TSS (--window=TSS)
3.3. Define a window relative to the TES (--window=TES)
3.4. Define a window relative to the gene body (--window=GeneBody)
3.5. Define a window relative to a column position (--window=<interger>)
4. Input file format3.2. Define a window relative to the TSS (--window=TSS)
3.3. Define a window relative to the TES (--window=TES)
3.4. Define a window relative to the gene body (--window=GeneBody)
3.5. Define a window relative to a column position (--window=<interger>)
5. Output file format (BED)
1. Overview
The GenomicIntervals2BED.py script converts a text-based input file into the standard BED file format. Additionally, it allows the user to define regions relative to coordinates within the file (relative to TSS, TES, peak maximum position, etc).
GenomicIntervals2BED.py is an important characteristic of SeqGI
as it confers flexibility
on the type of files that can be used. For instance columns can
be organized in
any order and separated by a range of common column delimiters (tab,
space, comma or semicolon).
2. Option summary
Usage: $ python GenomicIntervals2BED.py -f file [OPTIONS] -o outputname
The option --columns=2,3,4,5 specifies the column positions of the chromosome, start, end, strand information:
For example (--columns=2,3,4,5):
The option --cID=6 specifies the column position of the Ids.
When using this option, the ID column in the output BED file (column 4) is populated with the values of column 6 of the input file:
For example (--cID=6):
3.3. Define a window
relative to the TES (--window=TES)
Windows can also be defined relative to 3' end (transcription end
sites; TES),
For example, consider an asymmetrical TES window:

For example, a window flanking the TES would be defined as (--window=TES:-500:1000):
3.5. Define a window
relative to a column position (--window=<integer>)
The GenomicIntervals2BED.py script converts a text-based input file into the standard BED file format. Additionally, it allows the user to define regions relative to coordinates within the file (relative to TSS, TES, peak maximum position, etc).
- GenomicIntervals2BED.py allows you to convert an unstructured
text-base file into the standard BED format.
- Columns can be organized in
any order
- File delimiters can be tab, space, comma or semicolon
- Genomic regions of interest can be defined internally. Windows can be defined relative to:
- Gene related features (TSS, TES)
- Any column in the input file
2. Option summary
Usage: $ python GenomicIntervals2BED.py -f file [OPTIONS] -o outputname
| Options | Description |
| -h, --help | shows the full menu of available options |
| -f <file>, --fname=<file> | Complete path to input file. A text-base file with columns organized in any order. Mandatory fields include: chromosome, start, end (in any order). |
| -o <file>, --oname=<file> | Complete path to the output filename |
| -w <string>, --window=<string> |
[optional] Coordinates for window
relative to TSS, TES, gene body or any column in the input file.
Default is None. E.g. promoter coordinates centered at +-1kb of the TSS: --window=TSS:-1000:1000Other examples: --window=TES:0:2000A 500bp window flanking the coordinates on column2: --window=2:-500:500 |
| -t <string>, --sep=<string> | [optional] Separator of file (tab, comma, semicolon, space). Default is --sep=Tab |
| -c <string>, --columns=<string> |
[optional] chr,start,end,strand columns of file separated by commas. Strand is optional but needed if --wstrand. Default is --columns=1,2,3 |
| -i <string>, --cID=<string> |
[optional] Column nr of the ID column (e.g.: --cID=1). Default is None |
| -s, --wstrand | [optional]
Define window coordinates based on strand. This option takes into
account the orientation of the feature to define "upstream" and
"downstream" coordinates. Particularly important if defining asymmetrical windows. By default GenomeIntervals2BED does not take
strand information into account. |
| --onebased |
Specify this option if your input file has 1-based start positions. For instance, the GFF format from Ensemble uses 1-based coordinates for both the start and the end positions. While UCSC formats (such as BED files, or other files downloaded from the Table Browser) are 0-based at the start and 1-based at the end positions. By default, this script uses start positions as if they were 0-based. |
3.1. Convert any (yes! ANY) text file to BED: default behaviour (--window=None)
To convert a text-based file containing
information organized in any order, to a BED format, you need to
specify the column positions where the information on the coordinates
(chromosome, start, end ), strand and row IDs can be found.
The option --columns=2,3,4 specifies the column positions of the chromosome, start, end information:
For example (--columns=2,3,4):
The option --columns=2,3,4 specifies the column positions of the chromosome, start, end information:
For example (--columns=2,3,4):
| $ head mm9_canonical_chr1.txt #name chrom txStart txEnd strand kgXref.refseq uc007aeu.1 chr1 3204562 3661579 - NM_001011874 uc007aex.2 chr1 4333587 4350395 - NM_011283 uc007aez.1 chr1 4481008 4486494 - NM_011441 uc007aff.2 chr1 4763278 4775807 - NM_001177658 uc007afh.1 chr1 4797973 4836816 + NM_008866 uc007afi.2 chr1 4847774 4887990 + NM_011541 uc007afl.2 chr1 4899656 5060366 - NM_001177795 uc007afn.1 chr1 5073253 5152630 + NM_133826 uc007afo.1 chr1 5578573 5592947 + NM_011011 $ python GenomeIntervals2BED.py -f mm9_canonical_chr1.txt --columns=2,3,4 -o results.txt GenomeIntervals2BED6.py$head results.txt chr1 3204562 3661579 None 0 * chr1 4333587 4350395 None 0 * chr1 4481008 4486494 None 0 * chr1 4763278 4775807 None 0 * chr1 4797973 4836816 None 0 * chr1 4847774 4887990 None 0 * chr1 4899656 5060366 None 0 * chr1 5073253 5152630 None 0 * chr1 5578573 5592947 None 0 * chr1 5903787 5907479 None 0 * |
The option --columns=2,3,4,5 specifies the column positions of the chromosome, start, end, strand information:
For example (--columns=2,3,4,5):
| $ python GenomeIntervals2BED.py -f
mm9_canonical_chr1.txt --columns=2,3,4,5 -o
results.txt $head results.txt chr1 3204562 3661579 None 0 - chr1 4333587 4350395 None 0 - chr1 4481008 4486494 None 0 - chr1 4763278 4775807 None 0 - chr1 4797973 4836816 None 0 + chr1 4847774 4887990 None 0 + chr1 4899656 5060366 None 0 - chr1 5073253 5152630 None 0 + chr1 5578573 5592947 None 0 + chr1 5903787 5907479 None 0 - |
The option --cID=6 specifies the column position of the Ids.
When using this option, the ID column in the output BED file (column 4) is populated with the values of column 6 of the input file:
For example (--cID=6):
| $ python GenomeIntervals2BED.py -f
mm9_canonical_chr1.txt --columns=2,3,4,5
--cID=6 -o results.txt $head results.txt chr1 3204562 3661579 NM_001011874 0 - chr1 4333587 4350395 NM_011283 0 - chr1 4481008 4486494 NM_011441 0 - chr1 4763278 4775807 NM_001177658 0 - chr1 4797973 4836816 NM_008866 0 + chr1 4847774 4887990 NM_011541 0 + chr1 4899656 5060366 NM_001177795 0 - chr1 5073253 5152630 NM_133826 0 + chr1 5578573 5592947 NM_011011 0 + chr1 5903787 5907479 NM_010342 0 - |
3.2. Define a window relative to the TSS (--window=TSS)
A symmetrical TSS window centered at the
TSS would be defined as:

An asymmetrically TSS window would be defined as:

For example, 1kb window flanking the TSS would be defined as (--window=TSS:-1000:1000):
Note:
Windows defined relative to TSS, TES or the gene body require that the input file contains the strand information for each feature. The strand information is necessary to specify the upstream and downstream coordinates.
If strand information is not provided in the --columns option (i.e. chr,start,end,strand), GenomeIntervals2BED.py will give an error:

An asymmetrically TSS window would be defined as:

For example, 1kb window flanking the TSS would be defined as (--window=TSS:-1000:1000):
| $ head mm9_canonical_chr1.txt #name chrom txStart txEnd strand kgXref.refseq uc007aeu.1 chr1 3204562 3661579 - NM_001011874 uc007aex.2 chr1 4333587 4350395 - NM_011283 uc007aez.1 chr1 4481008 4486494 - NM_011441 uc007aff.2 chr1 4763278 4775807 - NM_001177658 uc007afh.1 chr1 4797973 4836816 + NM_008866 uc007afi.2 chr1 4847774 4887990 + NM_011541 uc007afl.2 chr1 4899656 5060366 - NM_001177795 uc007afn.1 chr1 5073253 5152630 + NM_133826 uc007afo.1 chr1 5578573 5592947 + NM_011011 $ python GenomeIntervals2BED.py -f mm9_canonical_chr1.txt --columns=2,3,4,5 --cID=6 --window=TSS:-1000:1000 -o results.txt GenomeIntervals2BED6.py Created file: results.txt$head results.txt chr1 3660579 3662579 NM_001011874 0 - chr1 4349395 4351395 NM_011283 0 - chr1 4485494 4487494 NM_011441 0 - chr1 4774807 4776807 NM_001177658 0 - chr1 4796973 4798973 NM_008866 0 + chr1 4846774 4848774 NM_011541 0 + chr1 5059366 5061366 NM_001177795 0 - chr1 5072253 5074253 NM_133826 0 + chr1 5577573 5579573 NM_011011 0 + chr1 5906479 5908479 NM_010342 0 - |
Note:
Windows defined relative to TSS, TES or the gene body require that the input file contains the strand information for each feature. The strand information is necessary to specify the upstream and downstream coordinates.
If strand information is not provided in the --columns option (i.e. chr,start,end,strand), GenomeIntervals2BED.py will give an error:
$ python GenomeIntervals2BED.py -f
mm9_canonical_chr1.txt --columns=2,3,4
--cID=6 --window=TSS:-1000:1000 -o
results.txt ERROR: --columns=2,3,4. Strand is needed to define windows based on strand |
Strand is needed to define TSS, TES and
GeneBody windows
- Windows defined relative to TSS, TES or the Gene body require
that the input file contains the strand information for each feature.
- Upstream and downstream coordinates are always defined based on strand. This means that, for instance the upstream interval will be on the right of a negative stranded feature, and on the left of a positive stranded feature.
- If strand is different than "+" or "-" the upstream and
downstream coordinates cannot be computed and the feature will be
skipped.
3.3. Define a window
relative to the TES (--window=TES)
Windows can also be defined relative to 3' end (transcription end
sites; TES),For example, consider an asymmetrical TES window:

For example, a window flanking the TES would be defined as (--window=TES:-500:1000):
| $ head mm9_canonical_chr1.txt #name chrom txStart txEnd strand kgXref.refseq uc007aeu.1 chr1 3204562 3661579 - NM_001011874 uc007aex.2 chr1 4333587 4350395 - NM_011283 uc007aez.1 chr1 4481008 4486494 - NM_011441 uc007aff.2 chr1 4763278 4775807 - NM_001177658 uc007afh.1 chr1 4797973 4836816 + NM_008866 uc007afi.2 chr1 4847774 4887990 + NM_011541 uc007afl.2 chr1 4899656 5060366 - NM_001177795 uc007afn.1 chr1 5073253 5152630 + NM_133826 uc007afo.1 chr1 5578573 5592947 + NM_011011 $ python GenomeIntervals2BED.py -f mm9_canonical_chr1.txt --columns=2,3,4,5 --cID=6 --window=TES:-500:1000 --wstrand -o results.txt GenomeIntervals2BED6.py$head results.txt chr1 3203562 3205062 NM_001011874 0 - chr1 4332587 4334087 NM_011283 0 - chr1 4480008 4481508 NM_011441 0 - chr1 4762278 4763778 NM_001177658 0 - chr1 4836316 4837816 NM_008866 0 + chr1 4887490 4888990 NM_011541 0 + chr1 4898656 4900156 NM_001177795 0 - chr1 5152130 5153630 NM_133826 0 + chr1 5592447 5593947 NM_011011 0 + chr1 5902787 5904287 NM_010342 0 - |
Windows can also be defined relative to
the gene body.
A Gene body window is defined relative to both, TSS and TES. Using in this case the coordinates upstream the TSS and downstream the TES:

A Gene body window is defined relative to both, TSS and TES. Using in this case the coordinates upstream the TSS and downstream the TES:

For example, a window flanking the gene
body region would be defined as (--window=GeneBody:-500:1000):
| $ head mm9_canonical_chr1.txt #name chrom txStart txEnd strand kgXref.refseq uc007aeu.1 chr1 3204562 3661579 - NM_001011874 uc007aex.2 chr1 4333587 4350395 - NM_011283 uc007aez.1 chr1 4481008 4486494 - NM_011441 uc007aff.2 chr1 4763278 4775807 - NM_001177658 uc007afh.1 chr1 4797973 4836816 + NM_008866 uc007afi.2 chr1 4847774 4887990 + NM_011541 uc007afl.2 chr1 4899656 5060366 - NM_001177795 uc007afn.1 chr1 5073253 5152630 + NM_133826 uc007afo.1 chr1 5578573 5592947 + NM_011011 $ python GenomeIntervals2BED.py -f mm9_canonical_chr1.txt --columns=2,3,4,5 --cID=6 --window=TES:-500:1000 --wstrand -o results.txt GenomeIntervals2BED6.py$head results.txt chr1 3203562 3662079 NM_001011874 0 - chr1 4332587 4350895 NM_011283 0 - chr1 4480008 4486994 NM_011441 0 - chr1 4762278 4776307 NM_001177658 0 - chr1 4797473 4837816 NM_008866 0 + chr1 4847274 4888990 NM_011541 0 + chr1 4898656 5060866 NM_001177795 0 - chr1 5072753 5153630 NM_133826 0 + chr1 5578073 5593947 NM_011011 0 + chr1 5902787 5907979 NM_010342 0 - |
3.5. Define a window
relative to a column position (--window=<integer>)
In some cases you might be interested in
investigating the pattern of read distribution in a genomic interval
defined relative to a given starting coordinate. For instance, the
column position can encode peak maximum positions, SNPs, etc.
In this case, GenomeIntervals2BED.py allows you to define the window of interest relative to coordinates present in a given column of the input file.
For example, 2kb window flanking the coordinates in column 2 and with --wstrand:
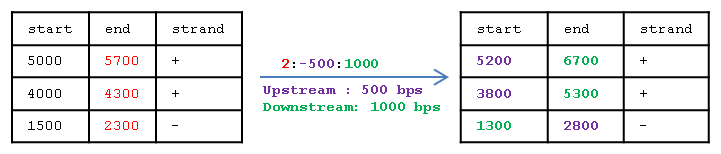
For example, a window flanking the TES would be defined as (--window=2:-500:1000):
Note:
Windows defined relative to the information in a particular column do not require that the input file to contain the strand information. Strand information is optional in this case.
In this case, GenomeIntervals2BED.py allows you to define the window of interest relative to coordinates present in a given column of the input file.
For example, 2kb window flanking the coordinates in column 2 and with --wstrand:

For example, a window flanking the TES would be defined as (--window=2:-500:1000):
| $ head mm9_canonical_chr1.txt #name chrom txStart txEnd strand kgXref.refseq uc007aeu.1 chr1 3204562 3661579 - NM_001011874 uc007aex.2 chr1 4333587 4350395 - NM_011283 uc007aez.1 chr1 4481008 4486494 - NM_011441 uc007aff.2 chr1 4763278 4775807 - NM_001177658 uc007afh.1 chr1 4797973 4836816 + NM_008866 uc007afi.2 chr1 4847774 4887990 + NM_011541 uc007afl.2 chr1 4899656 5060366 - NM_001177795 uc007afn.1 chr1 5073253 5152630 + NM_133826 uc007afo.1 chr1 5578573 5592947 + NM_011011 $ python GenomeIntervals2BED.py -f mm9_canonical_chr1.txt --columns=2,3,4,5 --cID=6 --window=3:-5:10 --wstrand -o results.txt GenomeIntervals2BED6.py$head results.txt chr1 3204552 3204567 NM_001011874 0 - chr1 4333577 4333592 NM_011283 0 - chr1 4480998 4481013 NM_011441 0 - chr1 4763268 4763283 NM_001177658 0 - chr1 4797968 4797983 NM_008866 0 + chr1 4847769 4847784 NM_011541 0 + chr1 4899646 4899661 NM_001177795 0 - chr1 5073248 5073263 NM_133826 0 + chr1 5578568 5578583 NM_011011 0 + chr1 5903777 5903792 NM_010342 0 - |
Note:
Windows defined relative to the information in a particular column do not require that the input file to contain the strand information. Strand information is optional in this case.
Strand is optional when defining windows
relative to column positions:
- If --wstrand is provided upstream
and downstream coordinates are defined based on strand. This means
that, for instance the upstream interval will be on the right of a
negative stranded feature, and on the left of a positive stranded
feature. Strand information is expected to be "+" or "-". Any other
value different than "+" or "-" will be skipped
- If --wstrand is not provided
then the features will be treated as "unstranded" and the (upstream,
downstream) coordinates will be defined as if the feature is in the
forward strand (left and right, respectively).
4. Input file
format
“Genomic Intervals” is a standard text-based format which contains information that can be organized in any order. File delimiters allowed include tab, space, comma or semicolon.
“Genomic Intervals” is a standard text-based format which contains information that can be organized in any order. File delimiters allowed include tab, space, comma or semicolon.
- Mandatory fields include: chromosome, start, end
- Optional fields include: strand, ID
- OGRe will skip any line that starts with "track", "browser" or the comment character "#".
- The strand field can only be "+" or "-".
5. Output file format (BED)
BED is tabular format also developed for use with the UCSC genome browser (see http://genome.ucsc.edu/FAQ/FAQformat#format1). The first three fields are mandatory and consist of chromosome, start, end. If "BED" is specified as the file format type, OGRe will make use of the first three fields. strand (position 6) and ID (position 4) will only be used if available. All the other fields will be ignored by OGRe.BED3 format: chromosome start end
BED4 format: chromosome start end ID
BED5 format: chromosome start end ID score
BED6 format: chromosome start end ID score strand
